Processing resources
This section will provide descriptions of the systems associate with BioKEM, for technical guides on how to use the systems, see How to use Blanca.
Petalibrary
Petalibrary is CU’s Research Computing (RC) files storage system. This service can be used to safely and securely store 100’s of TBs of data for a modest cost. This system is well backed up, so you don’t have to worry about losing valuable data. The other important aspect of this service is that you can securely mount it to other RC resources like Blanca for super fast file transfer.
You will need to request and allocation for you lab and have RC create a biokem-deposit folder that can be used to deposit images taken from the Krios. See https://www.colorado.edu/rc/resources/petalibrary for more information and email rc-help@colorado.edu to request and allocation. We recommend requesting an allocation of about ~20TB/actively processing dataset. This service could also be a good long term storage option for already processed datasets.

Blanca-biokem
Blanca is RC’s condo computing cluster. This cluster allows users to purchase their own computing nodes that RC will then house, setup, and manage. Users have priority on the nodes they own, but have access to use other users nodes when they are available. The BioKEM facility owns two types of nodes: one reserved for on the fly motion correction and the other for users of the facility to process EM data. The physical space we are allotted can house up to 10 processing nodes in addition to the on the fly node. These nodes are purchased by individual labs, but are then shared by all members of the BioKEM facility. Each node will consist of a number of GPUs (typically 4) and CPUs. All jobs ran in the cluster are submitted by a workload manager called SLURM.
Blanca-viz
In addition to the processing, login and compile nodes on Blanca, there are two GPU-accelerated visualization nodes that allow users to run graphical user interfaces, like ChimeraX. This is done through a web-based portal called CURC Open OnDemand and is how even command line based computing should be done on Blanca.
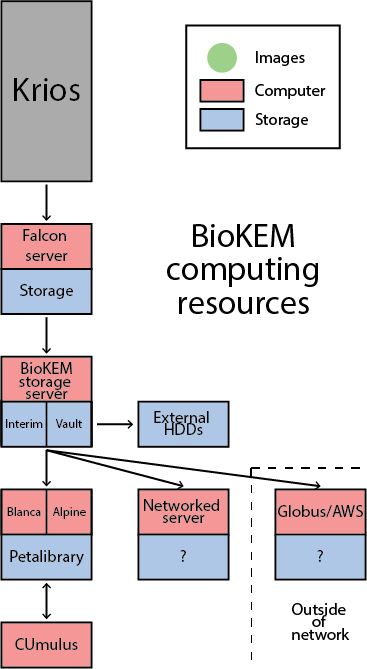
CUmulus
In addition to the Blanca computing cluster, users from CU also have the option to access CUmulus. CUmulus is CU’s cloud computing service that allows users to create virtual machines to fill specific roles that Blanca cannot perform for security or job limitation reasons. Two specific uses of this resource are: hosting a persistent job such as a CryoSPARC master instance or setting up a webpage. Clonable VMs to accomplish these tasks will be available to BioKEM users soon.
SLURM
SLURM is the management system many computing clusters, including Blanca use to run jobs. It allows multiple users to use the same computing resources by distributing jobs across resources and scheduling jobs to start when resources become available. The main benefits to us of using SLURM are:
Maximize utilization of nodes by running 24/7
Avoid single users from hogging resources
Create easy to replicate workflows
Allows us to use non-BioKEM nodes when they are available
All of these will bring the cost of high performance computing per lab down drastically, while working within a consistent compute environment that should be easier for the community to troubleshoot than working in individual labs.
SBGrid
To manage all of the software necessary for processing EM data, we are using a software manager called SBGrid. This service allows us to maintain multiple versions of software, as well as easily install and update new software.
Labs interested in using the suite of ~400 programs must purchase a lab specific license from SBGrid, we will then grant lab members access to these applications.